|
|
Post by camv35s on Sept 17, 2009 20:23:59 GMT -5
Hi Kammy just a short quote. (Up till now all technologies were CON-TROLLABLE.ELECTRICITY,even nuclear power can be turned off. GM is the first irreversible technology in human history.When a GMO is released it is out of our control;we have no means to call it back. We can insert a transgene, but we cannot take the released transgene out. Since GMOs are self replicating, releasing them might have dire consequences for human and animal health and for the environment and can change Evolution.) Professor Susan Bardocz biochemist and nutritionist. The same scientist that created this PLAGUE are the only ones that could hope to remedy it . Regards Cam
|
|
|
|
Post by kammy on Sept 17, 2009 22:06:09 GMT -5
That may be true, Cam, but we have to be optimistic? We're hearing that we can live a long time with this illness and into a rip old age, we have to believe that somebody in the medical or science community is going to figure out a way to alleviate a symptom or two so that we can know a better quality of life? We're fighters by nature, we're not just going to stand by while this disease goes unchecked and possibly ravishes the entire planet? No... somebody do something! lol The Lord knows we're trying...
|
|
|
|
Post by camv35s on Sept 18, 2009 0:26:15 GMT -5
Hi kammy I am optimistic but we cant ask our family doctor to compare amino acid sequences to known allergens .this puzzle will be solved in a bio research lab.I dont even like the name Morgellons,I believe it has hurt all of us. They associate us with dillusions of parasitosis.I call it what it is ,Transgenic toxic protien disease, caused by horizontal gene transfer of gut bacteria. and we all express different symptoms because our genome is 1 in a billion maybe, thanks for the response best regards cam.
|
|
|
|
Post by camv35s on Sept 18, 2009 0:49:10 GMT -5
Hi Kammy here could be a useful tool This overview includes a description of the discovery of RNA interference and “RNA silencing” (a more general term for the variety of recently discovered sequence-specific cellular responses to RNA), a summary of current thinking on the mechanism of RNAi, and various approaches to RNAi experiments. The material presented in this overview, when not explicitly referenced, is covered in one of several recent reviews [1–7].
RNA Interference and RNA Silencing
RNA interference (RNAi) is the process of mRNA degradation that is induced by double-stranded RNA in a sequence-specific manner. RNAi has been observed in all eukaryotes, from yeast to mammals. The power and utility of RNAi for specifically silencing the expression of any gene for which sequence is available has driven its incredibly rapid adoption as a tool for reverse genetics in eukaryotic systems.
The RNAi pathway is thought to be an ancient mechanism for protecting the host and its genome against viruses and rogue genetic elements that use double-stranded RNA (dsRNA) in their life cycles. RNAi is now recognized to be but one of a larger set of sequence-specific cellular responses to RNA, collectively called RNA silencing. These responses have been shown to play a role not only in mRNA and dsRNA stability/degradation, but also in regulation of translation, transcription, chromatin structure, and genome integrity. In plants and animals, RNA silencing has been adapted to play a critical role in regulation of cell growth and differentiation using a class of small RNAs called microRNAs (miRNAs).
In all RNA silencing pathways, dsRNA is processed to a 21–30 nucleotide-long RNA, which then functions as a component of a “silencing complex” to specifically repress expression or function of a target gene or genomic region (Figure 1). Specifically, in the RNAi pathway, dsRNA is processed to short interfering RNA (siRNA): 21–25 bp dsRNA with dinucleotide 3' overhangs. One strand of the siRNA (the guide strand) is then assembled into an RNA-induced silencing complex (RISC) that cleaves the target mRNA. The siRNA is thought to provide target specificity to RISC through base pairing of the guide strand with the target mRNA.
Figure 1. General Steps and Methods of RNA Silencing. In all RNA silencing pathways, double-stranded RNA (dsRNA) is processed to a small RNA which is assembled with RISC into a silencing complex that specifically represses expression or function of a target gene or genomic region by cleaving the corresponding mRNA.
When RNAi is artificially induced for reverse genetics, scientists use one of several forms of dsRNA to trigger the pathway. This topic is discussed in detail in subsequent sections of this overview.
The small RNAs that provide target specificity to the silencing machinery—short interfering RNAs (siRNAs), repeat-associated siRNAs (rasiRNAs), and microRNAs (miRNAs)—can be distinguished by their origin. siRNAs are processed from dsRNA precursors made up of two distinct strands of perfectly base-paired RNA, while miRNAs originate from a single, long transcript that forms imperfectly base-paired hairpin structures. siRNAs were originally identified as intermediates in the RNAi pathway after induction by exogenous dsRNA; however, endogenous sources of siRNAs have now been recognized. Many of these endogenous siRNAs are derived from repetitive sequences within the genome, hence the term repeat-associated siRNAs, or rasiRNAs. miRNAs were discovered through their critical roles in development and cellular regulation, and represent a large class of evolutionarily conserved RNAs. miRNAs have always been recognized as being of endogenous origin. However, synthetic precursors and inhibitors of miRNAs are used to understand and exploit the various functions of this important class of small RNA.
Although small RNAs differ in their origins, some of the proteins involved in their production and function in silencing complexes are closely related, and in some cases are identical. These include the most well characterized components of the silencing machinery, Dicers and Argonautes. Dicer produces small RNAs from longer precursors and plays a key role in assembling the guide strand of the small RNA with a pathway-specific complex of silencing proteins. Argonaute is a core component of all silencing complexes characterized so far and associates with the small RNA in the silencing complex. In the case of RNAi, Argonaute has been shown to be the catalytic engine of mRNA cleavage.
Discovery of RNAi
RNA interference was first observed in petunias, when Napoli et al. (1990) [8] discovered that introduction of a pigment-producing gene under control of a powerful promoter suppressed expression of both the introduced gene and the homologous endogenous gene, a phenomenon they called “cosuppression.” Cosuppression was subsequently found to occur in many species of plants and fungi (where it was called “quelling”) and to occur at the post-transcriptional level [8–10].
Such post-transcriptional gene silencing (PTGS) was shown to be mediated by a diffusible, trans-acting molecule in both Neurospora and plants [9, 11]. The crucial 1998 discovery by Fire et al., that injection of dsRNA—a mixture of both sense and antisense strands of the target mRNA, rather than either strand alone—into gonads of the nematode Caenorhabditis elegans resulted in extremely potent silencing unequivocably identified dsRNA as the inducer of RNA interference [12].
Short (~25 nt), antisense RNAs were first implicated in PTGS in plants [13]. Further work in Drosophila-derived in vitro systems showed that long dsRNA was processed to short interfering RNAs (siRNAs), 21–23 nt long, that could mediate target mRNA cleavage in the region corresponding to the introduced siRNA [14–16]. Current thinking about the mechanism of RNAi and components of the RNA-induced silencing complex is described more fully in the text.
In C. elegans, the demonstration that silencing could be induced by simply feeding nematodes with bacteria that had been engineered to express dsRNA homologous to the target gene, or by soaking worms in such dsRNA, drove rapid adoption of RNAi as a technology for reverse genetics at the organismal level [17–19]. Likewise, technology was quickly developed for inducing RNAi in Drosophila cell culture by bathing or transfection with dsRNA (see Armknecht et al. 2005 [20]).
Efforts to use RNAi in mammalian cells were hampered at first because of a nonspecific, interferon-mediated response to dsRNA longer than 30 bp that is seen in most mammalian cell lines. With the demonstration that 21–22 nt siRNAs, either chemically synthesized or expressed from a plasmid vector, could efficiently induce RNAi in mammalian cells without inducing the inteferon response, the door was opened for development of RNAi tools in mammalian systems [21, 22].
While some researchers were studying the phenomenon of RNAi and working to exploit its use as a tool for reverse genetics, others were trying to understand a distinct but related set of small RNAs—microRNAs (miRNAs). The first miRNAs to be discovered, lin-4 and let-7, were identified through loss-of-function mutations affecting control of postembryonic development in C. elegans [23, 24]. Other miRNAs were soon identified in C. elegans, Drosophila, and mouse by combinations of forward genetics, cDNA cloning, bioinformatics, and reverse genetics. By 2002, miRNAs were firmly established as a large class of conserved regulatory molecules in animals and plants [25–29]. miRNAs have been shown to play a role in developmental timing, cell death, cell proliferation, and oncogenesis [30–32]. This class of small RNA may represent 2–3% of the total number of genes in humans [4, 33], and estimates of miRNA target-binding sites indicate that miRNAs may play a role in regulating as many as 30% of mammalian genes [34].
A formal connection between the phenomenon of RNA interference and miRNAs was made when the same enzyme, Dicer, was shown to process both long dsRNA into siRNAs and cytoplasmic miRNA precursors (pre-miRNAs) into mature miRNAs [35–38]. Other components of the RNAi and miRNA silencing pathways have been shown to be closely related, most prominently Argonaute, which is a key component of the RNA-induced Silencing Complex (RISC). It now appears that miRNAs function in a silencing complex that is similar, if not identical, to RISC to regulate expression of target genes either through cleavage of mRNA or translational repression: if the miRNA exhibits perfect complementarity to its target mRNA, the mRNA is cleaved (typically in plants); if there is only partial complementary, translational repression occurs (typically in animals). The further discovery of endogenous small RNAs, distinct from miRNAs, that function in transcriptional silencing and genome stability has driven the adoption of the more general terms “RNA silencing” and “small RNAs” to describe the collection of related silencing pathways and their RNA guides. The term “RNA interference” (RNAi) continues to describe mRNA cleavage that is induced by a dsRNA trigger.
Small RNAs: siRNAs, rasiRNAs, and miRNAs
siRNAs and rasiRNAs
Short interfering RNAs (siRNAs) were originally identified as intermediates in the RNA interference pathway. siRNAs are 21–25 bp dsRNA with dinucleotide 3' overhangs that are formed in the cell from longer dsRNA molecules. The fully assembled RNA-induced silencing complex (RISC) contains only one strand of the siRNA, the guide strand. The guide strand is thought to provide target specificity for RISC-mediated cleavage through perfect base pairing with the mRNA target.
Long dsRNA (several hundred base pairs) can be used to artificially induce the RNAi pathway in fungi, plants, insects, and worms. In mammalian cells, dsRNA >30 bp induces a nonspecific, antiviral interferon response. However, synthetic siRNAs, ~21 bp with dinucleotide 3' overhangs, are potent triggers of RNAi in mammalian cells.
Although siRNAs are typically thought of as originating from exogenous precursors or triggers, it is perhaps not surprising that siRNAs generated from endogenous precursors have been identified. These endogenous siRNAs have been found to function in maintenance of chromatin structure and genome integrity as well as post-transcriptional gene silencing.
Endogenous siRNAs have been identified in plants, fungi, and animals. These siRNAs are derived in vivo from perfectly base-paired dsRNA precursors comprised of two distinct RNA strands. In many cases, endogenous siRNAs originate from repetitive elements within the genome, such as heterochromatic regions at centromeres and telomeres, and are therefore known as repeat-associated siRNAs (rasiRNAs). rasiRNAs appear to function, through an RNA-induced transcriptional silencing (RITS) complex, in maintaining the heterochromatic, and hence transcriptionally repressed, state of the region that encodes the rasiRNA. In Drosophila, rasiRNAs encoded by the Y-encoded Su(Ste) locus have also been implicated in trans regulation of the X-linked Stellate (Ste) locus, but it is not clear if silencing is at the transcriptional or post-transcriptional level [39]. In the protozoan Tetrahymena, endogenous siRNAs have also been shown to play a role in specifying sequences that are eliminated during macronucleus formation after sexual conjugation (Sontheimer and Carthew 2005 [4] and references within).
Endogenous nonrepetitive siRNAs have also been identified in the model plant Arabidopsis as well as in Caenorhabditis elegans [40, 41]. In some cases these endogenous siRNAs are complementary to longer, unique, coding RNAs. In Arabidopsis, nonrepetitive siRNAs have been implicated in regulation of transcript abundance through mRNA cleavage [41], indicating an RNAi mechanism similar to that triggered by exogenous siRNAs.
miRNAs
MicroRNAs (miRNAs) are a large class of evolutionarily conserved RNAs found in plants and animals. These small RNAs have been shown to play critical roles in developmental timing, hematopoietic cell differentiation, cell death, cell proliferation, and oncogenesis [30–32]. miRNAs may represent 2–3% of the total number of genes in humans [4, 33], and estimates of the number of miRNA target binding sites indicate that miRNAs may play a role in regulating as many as 30% of mammalian genes [34].
miRNAs are 19–23 nt single-stranded RNAs, originating from single-stranded precursor transcripts that are characterized by imperfectly base-paired hairpins. miRNAs function in a silencing complex that is similar, if not identical, to RISC.
In plants, miRNAs resemble siRNAs in that they exhibit perfect complementarity to binding sites in target mRNA and appear to silence gene expression through mRNA cleavage. Animal miRNAs typically exhibit partial complementarity to target binding sites in the 3' untranslated region (UTR) of target mRNA and repress translation without mRNA cleavage. It originally appeared that the process of miRNA gene silencing in animals involved reduced steady-state protein levels for the targeted gene without a reduction in the corresponding levels of mRNA; however, examples to the contrary have challenged this idea [4, 6]. miRNAs have also been implicated in mRNA turnover pathways in Drosophila and humans that use AU-rich elements (ARE), which are found in the 3' UTRs of many short-lived transcripts. miRNAs are also implicated in transcriptional silencing via hypermethylation in Arabidopsis [4].
Biogenesis of miRNAs and Endogenous siRNAs
miRNAs are synthesized in the nucleus as long (up to 1000 nt) RNA polymerase II transcripts, called pri-miRNAs, that are characterized by imperfect hairpin structures. Many pri-miRNAs arise from intergenic regions, or are in antisense orientation to known genes, indicating independent transcription units. Other genes for miRNAs are found in intronic regions and could be transcribed as part of the primary transcript for the corresponding gene [42]. A dsRNA-specific endonuclease, Drosha, in conjunction with a dsRNA-binding protein, called Pasha in Drosophila and DGCR8 in humans, processes the pri-miRNA into hairpin RNAs 70–100 nt in length, called pre-miRNAs. Pre-miRNAs are transported to the cytoplasm via an Exportin-5 dependent mechanism. There Dicer works with a dsRNA-binding partner, Loqs in Drosophila and TRBP in humans, to process the pre-miRNA into mature, single-stranded miRNA and load it into an RNA silencing complex that is similar to RISC. Loqs and TRBP are functionally analogous to the dsRNA-binding protein R2D2 which partners with Dicer to process and assemble siRNAs into RISC.
Endogenous dsRNA precursors for siRNAs could, in principle, arise from bidirectional transcription of genomic regions encoding siRNAs. However, in C. elegans, Arabidopsis, and fungi, silencing requires an RNA-dependent RNA polymerase (RdRP), raising the possibility that RdRP activity generates dsRNA from single-stranded RNA transcripts. Synthesis of dsRNA using the target mRNA itself as a template (using siRNA as a primer, or via de novo synthesis) could explain the phenomenon of “spreading” of RNAi, the production of siRNAs encoded by the target gene but not by the trigger RNA. Spreading is associated with “systemic silencing” seen in C. elegans and plants, in which silencing is inherited or is spread to distant parts of the organism. RdRP enzymes have not been found in Drosophila and mammals, and the spreading and systemic silencing phenomena are not seen in these organisms.
Many questions remain about endogenous siRNA production and function. How the cell distinguishes between normal mRNA and aberrant single-stranded RNA that is processed to dsRNA triggers for RNA silencing is unknown. Endogenous siRNA precursors are presumably transported to the cytoplasm before Dicer processing; the mechanism by which this happens is unknown, but is presumed to be similar if not identical to Exportin-5 mediated transport of pre-miRNAs. The biogenesis of endogenous siRNAs remains to be more clearly defined in all systems.
Definitions
Argonaute
Argonaute proteins are found in all RISC and miRNA-containing ribonucleoprotein complexes and have been shown to be the catalytic site of mRNA cleavage in RISC [47]. Argonautes are ~100 kDa proteins characterized by conserved domains called PAZ and PIWI. Argonautes can be organized by sequence into two subfamilies, Ago and Piwi, based on a higher degree of homology to either Arabidopsis AGO1 or to Drosophila Piwi. The Ago group functions in RNAi and miRNA silencing pathways.
Cosuppression
Silencing of an endogenous gene caused by the introduction of a trans-gene or infection by a virus. This term, which can refer to silencing at the post-transcriptional (PTGS; post-transcriptional gene silencing) or transcriptional (TGS; transcriptional gene silencing) level, is used primarily by researchers working with plants. In Neurospora crassa, this phenomenon is known as quelling.
Dicer
A ~200 kDa multidomain, RNase III family enzyme that functions in processing dsRNA to siRNA and assembly of the guide strand into RISC. Dicer progressively cleaves dsRNA at 21–25 bp intervals to generate siRNAs with 2-nt 3' overhangs and phosphorylated 5' ends. The predicted structure of Dicer includes an ATPase/RNA helicase domain, a conserved PAZ domain that is shared with Argonaute, two catalytic RNase III domains, and a C-terminal dsRNA binding domain (dsRBD).
dsRNA
Double-stranded RNA
Guide Strand
The strand of the siRNA that is assembled with RISC and provides sequence specificity for target mRNA cleavage. The guide strand is in the anti-sense orientation with respect to the mRNA.
miRNA
MicroRNA, a large class of evolutionarily conserved, noncoding, RNA originating from longer transcripts characterized by imperfect hairpin structures. miRNAs are 19–23 nt RNAs processed from pre-miRNA precursors by Dicer, the same enzyme that processes siRNAs.
Passenger Strand
The strand of the siRNA that dissociates from the siRNA during assembly with RISC.
PAZ Domain
Conserved domain found in both Dicer and Argonaute. It is thought that the 3' end of the guide strand of an siRNA is in contact with the PAZ in RISC.
PIWI Domain
Conserved domain found in Argonaute and thought to be the catalytic site for mRNA cleavage. It is thought that the 5' end of the guide strand of the siRNA contacts the PIWI domain in RISC.
Post-transcriptional Gene Silencing (PTGS)
Silencing of an endogenous gene caused by the introduction of a homologous dsRNA, trans-gene or virus. In PTGS, the transcript of the silenced gene is synthesized but does not accumulate because it is rapidly degraded. This is a more general term than RNAi, since it can be triggered by several different means. best regards Cam.
R2D2
A small, dsRNA binding protein that works with Dicer in assembly of siRNA with RISC.
rasiRNA
Repeat-associated siRNA, an endogenous form of siRNA that originates from repetitive elements within the genome.
RdRPs
RNA-dependent RNA Polymerases. RdRPs may play a role in amplifying RNA triggers for silencing. Although they are not found in insects and mammals, they are present in other eukaryotes that have RNA silencing pathways.
RISC
RNA-induced silencing complex. A nuclease complex, composed of proteins and siRNA, that targets and cleaves endogenous mRNAs complementary to the siRNA within the complex.
RITS
RNA-induced transcriptional silencing complex. A complex of proteins and rasiRNA that inhibits transcription of target DNA through heterochromatin formation. RITS contains at least one protein in common with RISC, Argonaute [49].
RNAi, RNA Interference
A process of mRNA cleavage and degradation that is induced by double-stranded RNA in a sequence-specific manner.
RNA Silencing
The collective term for the pathways that use small RNAs as guides to specifically modify expression of targeted genes or genomic regions.
RNase III Family
A group of double-stranded RNA-specific endonucleases characterized by production of dsRNA fragments with 2–3-nt 3' overhangs and 5' phosphorylated ends, similar to those produced by Dicer.
Small RNA
A collective term for siRNA, rasiRNA, and miRNA
siRNA
Short interfering RNA, siRNAs are 21–25 bp dsRNA with dinucleotide 3' overhangs that are processed from longer dsRNA by Dicer in the RNA interference pathway. Introduction of synthetic siRNAs can induce RNA interference in mammalian cells. siRNAs can also originate from endogenous dsRNA precursors.
shRNA
Short hairpin RNA. shRNAs are used in plasmid- or vector-based approaches for supplying siRNAs to cells to produce stable gene silencing. A strong promoter is used to drive transcription of a target sequence designed to form hairpins and loops of variable length, which are then processed to siRNAs by the cellular RNAi machinery.
siRISC
A term for RISC assembled with the guide strand of an siRNA.
Home | Products | Technical Resources | What's New | About Us | Contact Us
Advanced Search | Site Map | Privacy | Trademarks/Legal | Web Feedback | Jobs
©2009 Applied Biosystems. All rights reserved.
|
|
|
|
Post by fritolay66 on Sept 18, 2009 7:27:26 GMT -5
Chemically synthesized?
Expressed from a plasmid vector?
Without an interferon response?
I posted the lab results posted from the
MRF common to those of Morgellons. I do not see an
interferon result?
Development of what tools in mammals?
Proteins in function and production in silencing complexes are closely related or identical?
This screams at me.
Screaming again.
Precursors or triggers?
Chromatin structure, genome integrity, p-t gene silencing?
Questions remain as usual. They don't know how it differientates between the normal and abberant BUT the door was opened for development of RNAi tools in mammalian systems.
I am unfamilar with Exportin-5 mechanism. Anybody care to explain?
|
|
|
|
Post by Jill on Sept 18, 2009 9:56:25 GMT -5
as promised- more on the Cement cone- related to T parva: One patent- for Cement - for bonding human and animal tissue- and for vaccines- using T parva vectored by the Rhipicephalus appendiculatus tick: tinyurl.com/mv5l9lFair use Tissue cement proteins from Rhipicephalus appendiculatus Excerpt The present invention relates to tissue cement proteins produced by certain species of blood-feeding ectoparasites. These proteins and compositions comprising these proteins are particularly useful for the temporary or permanent bonding of animal tissues to each other or to other biomaterials. The present invention also relates to the use of tissue cement proteins in the production of vaccines that protect animals against the bite of blood-sucking ectoparasites and the transmission of viruses, bacteria and other pathogens by such ectoparasites. Excerpt: Description The present invention relates to tissue cement proteins produced by certain species of blood-feeding ectoparasites. These proteins and compositions comprising these proteins are particularly useful for the temporary or permanent bonding of animal tissues to each other or to other biomaterials. The present invention also relates to the use of tissue cement proteins in the production of vaccines that protect animals against the bite of blood-sucking ectoparasites and the transmission of viruses, bacteria and other pathogens by such ectoparasites Cement is produced by many blood-feeding ectoparasites, including certain species of Ixodid ticks Ixodid (hard) ticks are haematophagous parasites that attach themselves to a vertebrate host by means of a `cement cone`, a product of the type II and type III acini of the tick salivary glands (Kemp et al., 1982. Walker et al., 1985) (All documents referred to herein are listed at the end of the description.)The cement that forms the cone is a milky-white secretion that is injected into the skin of animals on which these parasites feed. The cement comprises a number of interacting protein and carbohydrate components. The cement spreads into the bite site and over the skin and, upon hardening, ensures that the mouthparts remain firmly anchored to the host during the feeding period, which typically lasts 4 to 8 days. The cement cone functions additionally as a gasket to prevent leakage of fluids from the bite site during feeding.
The tick cement cone is a layered structure, constructed from two major types of cement. The first type of cement is produced just minutes after establishing the bite site and hardens quickly to form a rigid `core` of the cone. A second type of cement is secreted later, about 24 hours after attachment, and hardens more slowly to form a more flexible `cortex`. In adult ticks, cement production typically continues until the 3rd or 4th day after attachment (Kemp et al., 1982; Sonenshine et al., 1991).The tick cement cone appears to be mainly proteinaceous, but also contains some carbohydrate and lipid. An early study found the amino-acid composition of whole cement in Boophilus microplus to be rich in glycine, leucine, serine and tyrosine (Kemp et al., 1982) However, the individual proteins comprising the tick cement are very poorly characterised Although the mobility of the component proteins has been show on SDS-PAGE gels, none have yet been purified The process by which the cement components harden is also not understood, although mechanisms similar to the tanning of cuticle and coagulation of haemolymph have been proposed (Kemp et al., 1982; Moorhouse and Tatchell, 1966). At present no direct scientific evidence has been produced to substantiate these theoretical mechanisms. It has been noted that the polypeptides that form the cement cortex appear to be similar to certain structural components of vertebrate skin, involvement of these vertebrate-like molecules may enable ticks to use host-derived enzymes during the cement hardening process, for example lysyl oxidases which cross-link collagen and elastin (Siegel, 1979); or transglutaminases, such as the coagulation factor XIIIa, which is induced during wound healing and cross-links fibronectin, fibrins and collagen (Ichinose et al., 1990). These enzymes may cross-link cortex polypeptides to the extracellular matrix proteins of the skin. Other enzymes such as phenoloxidases or peroxidases which catalyse the hardening of arthropod extracellular structures (Sugumaran et al., 1992) have been identified in R. appendiculatus salivary glands and are therefore likely to play a role in solidifying the cement cone.
The composition of tick cement appears to be similar amongst different Ixodid tick species For example, an antiserum raised against a 90 kD salivary protein of the brown ear tick, Rhipicephalus appendiculatus, has been shown to recognise polypeptides from the salivary glands and cement proteins of the American dog tick, Dermacenter variabilis, the lone star tick, Amblyomma americanum, and the brown dog tick, R sanguineus (Jaworski et al., 1992)All these tick species are extremely effective as transmitters of disease For example, R appendiculatus represents a major obstacle to livestock development in several sub-saharan regions It transmits the protozoan parasite Theileria parva which causes the usually fatal East Coast Fever This disease is often considered the most important disease of cattle (Norval et al., 1992a, Norval et al., 1992b). This tick is also the main vector of the virus causing Nairobi sheep disease, a disabling and often deadly disease in sheep and goats (Davies. 1988). R. appendiculatus and other tick pests also cause considerable damage to the skin, thereby affecting the leather industry. In an effort to combat parasite-transmitted diseases, unpurified cement components have been tested as inducers of host resistance (Brown et al., 1986; Shapiro et al., 1989), but reliable vaccines based on cement proteins have not been successfully developed. Cone proteins would appear to be a reasonable target for a vaccine since the formation of the cone is essential for the tick to attach to the host and feed. However, only some of the cone proteins are antigenic. There therefore exists a great need for an effective vaccine to combat diseases that are transmitted by blood-feeding ectoparasites. The elucidation of the components of tissue cement produced by these organisms would allow the rational design of such vaccines. Furthermore, these molecules would prove useful in medicine as components of tissue cement. Presently available tissue cements are of two types, both of which suffer from significant disadvantages. Acrylic-based glues are extremely strong, yet are also very toxic and can thus only be used in very small quantities in the body. The second type of tissue cement used is non-immunogenic but forms a much less strong bond. Consequently this type of cement is only useful in a small number of surgical procedures. There is thus a great need for a non-immunogenic tissue cement that is capable of bonding mammalian tissue with great strength. |
|
|
|
Post by kammy on Sept 19, 2009 1:24:33 GMT -5
The present invention also relates to the use of tissue cement proteins in the production of vaccines that protect animals against the bite of blood-sucking ectoparasites and the transmission of viruses, bacteria and other pathogens by such ectoparasites. Thank you Jill and Cam for your input. Cam, don't go away... that was quite informative, it's going to take some 'processing'. Good points, Frito... So, Jill, has Robert Smith proven or is finding that this tick tissue cement was used in the T. parva vaccine? Or, is he finding any kind of glue, cement, rubbery-like material, artificial skin chemicals, etc. in the samples? The theory is that by the cattle being inoculated with the T. parva vaccine and it contained this particular cement protein, and by us ingesting the meat, milk or cheese - is why 90 something plus percent? of people with Morgellons are showing the T. parva markers and causing this 'cement cone' characteristic as being a part of our lesions? |
|
|
|
Post by kammy on Sept 19, 2009 4:24:13 GMT -5
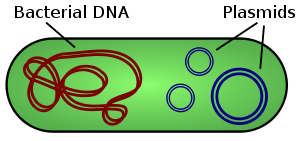
Ok, Cam, that kept me busy, I'm still studying it, and there's so much good information in your last post. From what I'm understanding, we would need to know the protein or RNA sequences of what's inside our disease that we all have in common in order to find a solution using RNA Interference and/or RNA Silencing? I believe this is what Dr. Wymore is currently doing using the 'shotgun method' that he spoke about in his last report? He's attempting to get us some RNA/DNA/protein? sequence numbers of what we all have in common, so that we can figure out what all is entailed with our disease. I hope he will publish that information soon? en.wikipedia.org/wiki/RNA-SeqI know there's protein sequencing software to do comparisons on the Internet, I'm not sure about RNA/DNA? When we get some sequence strings, then we can look to see what they match and go from there? ---------------------- (What's the web link address to this site article, Cam, so we can look at the References?) ---------------------- In the meantime, we might want to look at: "Cosuppression was subsequently found to occur in many species of plants and fungi (where it was called “quelling”) and to occur at the post-transcriptional level [8–10]. " Cosuppression or quelling, can they help us? ---------------------- "In C. elegans, the demonstration that silencing could be induced by simply feeding nematodes with bacteria that had been engineered to express dsRNA homologous to the target gene, or by soaking worms in such dsRNA, drove rapid adoption of RNAi as a technology for reverse genetics at the organismal level [17–19]. Likewise, technology was quickly developed for inducing RNAi in Drosophila cell culture by bathing or transfection with dsRNA (see Armknecht et al. 2005 [20])." When did this technology begin, how does it work? What company is producing it, what form does it come in when you purchase it? Patents? ---------------------- "Efforts to use RNAi in mammalian cells were hampered at first because of a nonspecific, interferon-mediated response to dsRNA longer than 30 bp that is seen in most mammalian cell lines. With the demonstration that 21–22 nt siRNAs, either chemically synthesized or expressed from a plasmid vector, could efficiently induce RNAi in mammalian cells without inducing the inteferon response, the door was opened for development of RNAi tools in mammalian systems [21, 22]." What year did this technology start? Plasmid vector: en.wikipedia.org/wiki/Plasmid_vector#Vectors"A plasmid is an extra-chromosomal DNA molecule separate from the chromosomal DNA which is capable of replicating independently of the chromosomal DNA.[1] In many cases, it is circular and double-stranded. Plasmids usually occur naturally in bacteria, but are sometimes found in eukaryotic organisms (e.g., the 2-micrometre-ring in Saccharomyces cerevisiae)." I believe that one of our spheres that is meeting the definition of a plasmid or plasmid vector, in the photographs we see a green 'ring' around the spheres. "Plasmids are considered transferable genetic elements, or "replicons", capable of autonomous replication within a suitable host. Plasmids can be found in all three major domains, Archea, Bacteria and Eukarya.[1] Similar to viruses, plasmids are not considered a form of "life" as it is currently defined.[6] Unlike viruses, plasmids are "naked" DNA and do not encode genes necessary to encase the genetic material for transfer to a new host. Plasmid host-to-host transfer requires direct, mechanical transfer by "conjugation" or changes in host gene expression allowing the intentional uptake of the genetic element by "transformation".[1] Microbial transformation with plasmid DNA is neither parasitic nor symbiotic in nature, since each implies the presence of an independent species living in a commensal or detrimental state with the host organism. Rather, plasmids provide a mechanism for horizontal gene transfer within a population of microbes and typically provide a selective advantage under a given environmental state. Plasmids may carry genes that provide resistance to naturally occurring antibiotics in a competitive environmental niche, or alternatively the proteins produced may act as toxins under similar circumstances. Plasmids also can provide bacteria with an ability to fix elemental nitrogen or to degrade calcitrant organic compounds which provide an advantage under conditions of nutrient deprivation.[1]" ---------------------- en.wikipedia.org/wiki/Transformation_%28genetics%29---------------------- We might look at Argonaute's role in stopping this system? ---------------------- "In Drosophila, rasiRNAs encoded by the Y-encoded Su(Ste) locus have also been implicated in trans regulation of the X-linked Stellate (Ste) locus, but it is not clear if silencing is at the transcriptional or post-transcriptional level [39]. In the protozoan Tetrahymena, endogenous siRNAs have also been shown to play a role in specifying sequences that are eliminated during macronucleus formation after sexual conjugation (Sontheimer and Carthew 2005 [4] and references within)." How does this statement pertain to us? ---------------------- "However, in C. elegans, Arabidopsis, and fungi, silencing requires an RNA-dependent RNA polymerase (RdRP), RdRP enzymes have not been found in Drosophila and mammals, and the spreading and systemic silencing phenomena are not seen in these organisms." RdRP enzymes? |
|
|
|
Post by kammy on Sept 19, 2009 5:26:47 GMT -5
|
|
|
|
Post by jeany on Sept 19, 2009 6:58:16 GMT -5
Hi folks, I did a little research on the Argonaute protein (Ago) and have found that a novel antibody called 2A8 was discovered which has the capability to develop monoclonal antibodies against mammalian Ago proteins. Here's the link and a few excerpts: www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1986805
2A8, a novel anti-Ago monoclonal antibody--A novel monoclonal antibody against human Argonaute proteins reveals unexpected characteristics of miRNAs in human blood cellsArgonaute (Ago) proteins bind to microRNA (miRNAs) and short interfering RNAs (siRNAs) and form the core components of effector complexes that mediate miRNA and siRNA function. Currently, there is a paucity of reliable antibodies against mammalian Ago proteins, thus precluding studies of endogenous Ago proteins from tissues. Here we report the development of 2A8, a novel anti-Ago monoclonal antibody that recognizes human and mouse Ago proteins and efficiently immunoprecipitates miRNAs.MicroRNAs (miRNAs) are one of the most prevalent small (~22 nucleotide [nt]) regulatory RNA classes in animals (for reviews, see Nelson et al. 2003; Cao et al. 2006). MiRNAs exert their influence through complementary base-pairing with specific “target” mRNAs, leading in turn to degradation or translational repression of the targeted mRNA. MiRNAs have been shown to play important roles in cell fate determination, development, apoptosis, and other fundamental biological processes.Currently, there is a paucity of antibodies against mammalian Ago proteins, limiting biochemical studies of endogenous Ago proteins and precluding isolation of Ago-associated miRNAs from tissues. To surmount these obstacles, we have developed a monoclonal antibody against mammalian Ago proteins.
The antibody, which is termed 2A8, recognizes all four human and mouse Ago proteins, and coimmunoprecipitates miRNAs.
**A test to determine these proteins and antibodies is Western Blot. I also found this interesting: We have also confirmed that the ~70 kDa band corresponded to radixin by Western blots of 2A8 immunoprecipitates with anti-radixin antibody (P. Nelson and Z. Mourelatos, unpubl.). However, anti-radixin antibodies did not co-immunoprecipitate Ago proteins (P. Nelson and Z. Mourelatos, unpubl.). Radixin is an actin-binding protein with no known role in RNA regulation (Bretscher 1999; Hoeflich and Ikura 2004). Hence, there is an apparent cross-reaction between 2A8 and radixin.en.wikipedia.org/wiki/RadixinRadixin is a cytoskeletal protein that may be important in linking actin to the plasma membrane.**Is there a relationship between Ago Protein and Actin Protein? **Past research on the Baculovirus has revealed that the protein FALPE and p10 has the characteristic to form Actin filaments. In my blog entry below, you can read how I think this might interact in humans. morgellons2.wordpress.com/2009/09/12/formation-of-actin-filaments-in-mammalian-cells-baculovirus-protein-falpe-and-p10/www.pubmedcentral.nih.gov/articlerender.fcgi?artid=113992Another aspect: excerpt: 'We show here that baculovirus can indeed infect nondividing mammalian cells through a mechanism apparently identical to that found in insect cells.' 'This suggests a mechanism of DNA transport similar to HSV.' **This kept me thinking...If we all have this Ago Protein and the Baculovirus introduces FALPE and P10 proteins, can this interfere with the Actin protein which in the other hand is responsible of filamentous tissue building in the cytoskeleton? And...Baculovirus has a similar DNA transport mechanism as Herpes? Is this the connection? Does the Baculovirus 'need' the Herpes Virus to infect us or invade our bodies? and these viruses interact with the above mentioned proteins which cause our symptoms...the filaments...our disease? hmm..just thinking here.. Jeany |
|
|
|
Post by jeany on Sept 19, 2009 11:30:26 GMT -5
www.ncbi.nlm.nih.gov/pubmed/9891781?dopt=AbstractPlus&holding=f1000,f1000m,isrctn Many pathogens actively exploit the actin cytoskeleton during infection. This exploitation may take place during entry into mammalian cells after engagement of a receptor and/or as series of signaling events culminating in the engulfment of the microorganism. Although actin rearrangements are a common feature of most internalization events (e.g. entry of Listeria, Salmonella, Shigella, Yersinia, Neisseria, and Bartonella), bacterial and other cellular factors involved in entry are specific to each bacterium. Another step during which pathogens harness the actin cytoskeleton takes place in the cytosol, within which some bacteria (Listeria, Shigella, Rickettsia) or viruses (vaccinia virus) are able to move. Movement is coupled to a polarized actin polymerization process, with the formation of characteristic actin tails. Increasing attention has focused on this phenomenon due to its striking similarity to cellular events occurring at the leading edge of locomoting cells. Thus pathogens are convenient systems in which to study actin cytoskeleton rearrangements in response to stimuli at the plasma membrane or inside cells.
|
|
|
|
Post by bannanny on Sept 19, 2009 19:21:19 GMT -5
What I don't understand is how I could even have Lyme. Is there a chance I've had it for years and just didn't know it? I know when I was a vet tech working in Tennessee, we had to shave a chow down completely. When I got home that night and took my jeans off to get in the shower, I had ticks all over my legs. That was back in the late 80's early 90's. Could I have been infected then and just never showed any symptoms?
|
|
|
|
Post by kammy on Sept 19, 2009 21:02:47 GMT -5
What I don't understand is how I could even have Lyme. Is there a chance I've had it for years and just didn't know it? I know when I was a vet tech working in Tennessee, we had to shave a chow down completely. When I got home that night and took my jeans off to get in the shower, I had ticks all over my legs. That was back in the late 80's early 90's. Could I have been infected then and just never showed any symptoms? I haven't studied Lymes too much, Banny, maybe someone here that has Lymes can better answer your questions? I could have it, I haven't been tested properly, yet. I believe that many of us could have Lymes and its co-infections and not know it. They have found where you don't necessarily have to have the 'bulls eye' rash or possibly not even be bit by a tick, that there might be other insects that are vectors now? Some of us can't even get a proper blood test to find out what's going on. Did you get the Western Blot test done, Banny, and do you know what your co-infections are, besides Morgellons? Jeany, that's good work, how do we stop or kill it? If we can stop one process, maybe it'll cause another to stop? I know some people have become symptom free - I wonder what they stumbled on? |
|
|
|
Post by bannanny on Sept 20, 2009 20:44:53 GMT -5
I had 2 tests done... the Elisia and the Igenex western blot. My doc said they were negative but he knows nothing about Lyme or how to read the tests. I finally posted my test results here and Kmarie and frito (I believe it was them) said the results showed I do have Lyme. I have all the symptoms on top of all the morg symptoms. I tend to think morgs might even be a mutant strain of Lyme. I don't know how else we could all be showing symptoms of Lyme as well as morgs if that weren't the case. Either that or Lyme was mixed with whatever morgs is... wonder if we'll ever know.
I have Fibromyalgia and CFS just like all of us supposedly do. I also have 2 spinal diseases, but I was diagnosed with all of those before I ever got morgs. The only thing I've gotten since morgs is I have granulomas in my lungs now.
As far as what the people did who are symptom free... it seems like the few I know of were all on completely different protocols from one another. So who knows what did it... maybe something in their genetic makeup?
hugs ~~ bannanny
|
|
|
|
Post by lilsissy on Sept 20, 2009 22:16:02 GMT -5
This is fascinating , the cement hey?
Wished I had more time right ow, just skimming but you guys are doing a great job.
Remember Yersinia pestis page from the N.I.H. had a morgellons paragraph on it.
Jen
|
|
|
|
Post by camv35s on Sept 20, 2009 23:23:13 GMT -5
We investigated the antiviral activity of olive leaf extract (OLE) preparations standardized by liquid chromatography-coupled mass spectrometry (LC-MS) against HIV-1 infection and replication. We find that OLE inhibits acute infection and cell-to-cell transmission of HIV-1 as assayed by syncytia formation using uninfected MT2 cells co-cultured with HIV-1-infected H9 T lymphocytes.
Most Recent
Health Care Articles
Physicians Finally Getting Help to Become Medical Homes
Obama Tort Reform Initiative Won't Get Us Far
"Health 2.0" vs. Consumer Apathy
Comparative Effectiveness: Putting Medicine Under a Microscope
Insurers Take On Botched Medical Work -- With Their Checkbooks
More »
OLE also inhibits HIV-1 replication as assayed by p24 expression in infected H9 cells. These anti-HIV effects of OLE are dose dependent, with EC(50)s of around 0.2 microg/ml. In the effective dose range, no cytotoxicity on uninfected target cells was detected. The therapeutic index of OLE is above 5000. To identify viral and host targets for OLE, we characterized gene expression profiles associated with HIV-1 infection and OLE treatment using cDNA microarrays. HIV-1 infection modulates the expression patterns of cellular genes involved in apoptosis, stress, cytokine, protein kinase C, and hedgehog signaling. HIV-1 infection up-regulates the expression of the heat-shock proteins hsp27 and hsp90, the DNA damage inducible transcript 1 gadd45, the p53-binding protein mdm2, and the hedgehog signal protein patched 1, while it down-regulates the expression of the anti-apoptotic BCL2-associated X protein Bax. Treatment with OLE reverses many of these HIV-1 infection-associated changes. Treatment of HIV-1-infected cells with OLE also up-regulates the expression of the apoptosis inhibitor proteins IAP1 and 2, as well as the calcium and protein kinase C pathway signaling molecules IL-2, IL-2Ralpha, and ornithine decarboxylase ODC 1.
This is part of my protocol if it has a positive effect on aids, a transgenic disease why not our problem. best regards Cam
|
|
|
|
Post by camv35s on Sept 20, 2009 23:44:23 GMT -5
Hi bananny MSM is a source of biological sulfur, which is a major component in many of the body's proteins, tissues, hormones and enzymes. Sulfur also plays a role in the detoxification of the liver. Because MSM can inhibit pain impulses, promote blood flow, and reduce inflammation of tissues. It has also been researched for use as a pain reliever and anti-inflammatory treatment. MSM has been researched for its ability to reduce pain associated with a long list of disorders. Some of these include:
Fibromyalgia
Back pain from herniated discs, and other causes
Headaches
Muscle soreness
Tendinitis
Bursitis
Carpal tunnel syndrome
Athletic strains and sprains
Cold sores
Inflammatory bowel disorders
Shingles
TMJ (temporal mandibular joint) pain
MSM has seen use in treating allergies and asthma.(1) It has also been used to ease constipation and bladder inflammation.(2),(3),(4) It may even reduce the inflammation associated with gingivitis when used as a mouthwash.(5)
Research suggests that MSM may have a variety of benefits for people with all types of arthritis. Benefits can include the reduction, or even elimination, of pain in some cases.(6) For osteoarthritis, studies suggest that MSM may inhibit the formation of scar tissue around joints and slow down degeneration of cartilage.(7),(8),(9)
When applied topically, MSM may also reduce scarring and provide benefit for people with scleroderma, a disorder that involves hardening of the skin.(10),(11) Additionally, MSM has shown promise in treating many of the symptoms of lupus.(12),(13)
Toxicities & Precautions
More information about Methyl Sulfonyl Methane (MSM)
Be sure to tell your pharmacist, doctor, or other health care providers about any dietary supplements you are taking. There may be a potential for interactions or side effects.
Introduction
Be sure to tell your pharmacist, doctor, or other health care providers about any dietary supplements you are taking. There may be a potential for interactions or side effects.
General
This dietary supplement is considered safe when used in accordance with proper dosing guidelines.
Side Effects
Occasional side effects reported with large doses of this dietary supplement include mild stomach upset, occasional headaches and more frequent bowel movements. It may be necessary to reduce the dose of this dietary supplement. Tell your doctor if these side effects become severe or do not go away.
Pregnancy/Breast-Feeding
To date, the medical literature has not reported any adverse effects related to fetal development during pregnancy or to infants who are breast-fed. Yet little is known about the use of this dietary supplement while pregnant or breast-feeding. Therefore, it is recommended that you inform your healthcare practitioner of any dietary supplements you are using while pregnant or breast-feeding.
Age Limitations
This supplement should not be used in children unless recommended by your physician.
Links
Content provided by NHI OnDemand, the trusted online source of current natural health & wellness information.
References
Jacob SW, et al. The Miracle of MSM: The Natural Solution for Pain. New York: G.P. Putnam's Sons; 1999:57-58.
Jacob SW, et al. The Miracle of MSM: The Natural Solution for Pain. New York: G.P. Putnam's Sons; 1999:57-58.
Jacob SW, et al. The Miracle of MSM: The Natural Solution for Pain. New York: G.P. Putnam's Sons; 1999:57-58.
View Abstract: Childs SJ. Dimethyl sulfone (DMSO2) in the Treatment of Interstitial Cystitis. Urol Clin North Am. Feb1994;21(1):85-88.
Jacob SW, et al. The Miracle of MSM: The Natural Solution for Pain. New York: G.P. Putnam's Sons; 1999:57-58.
Jacob SW, et al. The Miracle of MSM: The Natural Solution for Pain. New York: G.P. Putnam's Sons; 1999:57-58.
Jacob SW, et al. The Miracle of MSM: The Natural Solution for Pain. New York: G.P. Putnam's Sons; 1999:57-58.
View Abstract: Rizzo R. Calcium, Sulfur and Zinc Distribution in Normal and Arthritic Articular Equine Cartilage: A Syncrotron Radiation Induced X-ray Emission Study. Journal of Experimental Zoology. Sep1995;237(1):82-86.
Hess WC, Sullivan MS. Cystine Content in Fingernails in Arthritics. Journal of Bone Joint Surgery. 1935;16:185-88.
Jacob SW, et al. The Miracle of MSM: The Natural Solution for Pain. New York: G.P. Putnam's Sons; 1999:57-58.
Jacob SW, et al. The Miracle of MSM: The Natural Solution for Pain. New York: G.P. Putnam's Sons; 1999:57-58.
Jacob SW, et al. The Miracle of MSM: The Natural Solution for Pain. New York: G.P. Putnam's Sons; 1999:57-58.
View Abstract: Morton JI, Siegel BV. Effects of Oral Dimethyl Sulfoxide and Dimethyl Sulfone on Murine Autoimmune Lymphoproliferative Disease. Proceedings of the Society for Experimental Biology and Medicine. 1986;183:227-30.
This information is educational in context and is not to be used to diagnose, treat or cure any disease. Please consult your licensed health care practitioner before using this or any medical information.
©2000-2008 CCG, Inc. All Rights Reserved.Get Free Health Insurance Quotes!
Hi bananny this is the skinny on msm its great to gargle with for your teeth
|
|
|
|
Post by believer on Sept 21, 2009 1:13:09 GMT -5
May I ask what the best brand/form of MSM would be  Excellent research here too I may add, its so far over my head I feel short today, LOL !! Thank you ALL!!!!!! |
|
|
|
Post by camv35s on Sept 21, 2009 8:52:06 GMT -5
Hi believer, I use the NOW brand MSM Good quality Have used it for years ,Best regards CAM.
|
|
|
|
Post by fritolay66 on Sept 21, 2009 14:40:54 GMT -5
I am not sure whether you all would be interested in my Cats Claw post. I wasn't sure where to put it, so I put it in the supplements thread.
frito
|
|
 May 1, 2024 14:29:24 GMT -5
May 1, 2024 14:29:24 GMT -5
 May 1, 2024 14:29:24 GMT -5
May 1, 2024 14:29:24 GMT -5